Figure 1
Intestinal mTORC2 prevents dauer formation
1A
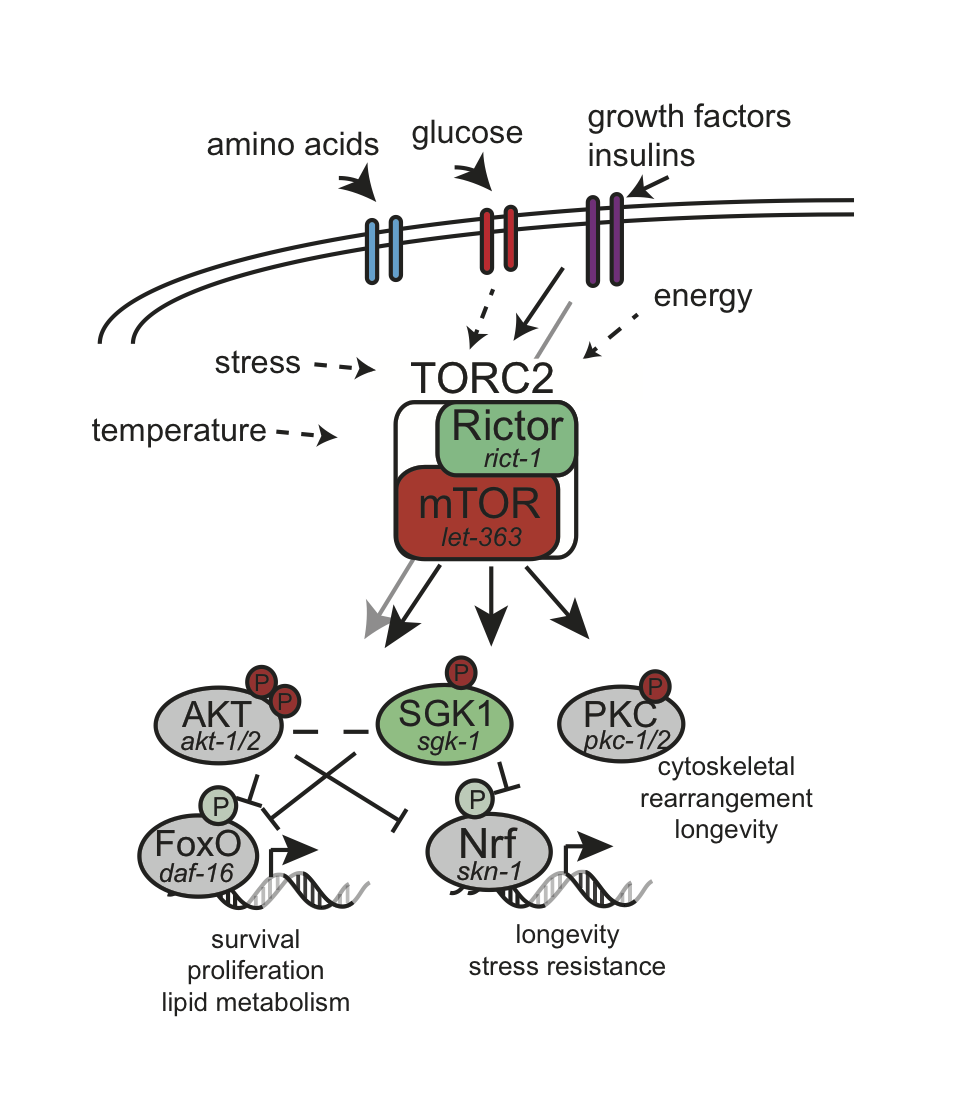
TOR complexes are involved in nutrient homostasis
Figure 1 The TORC2 signaling pathway in C. elegans (11, 53-55). Genes encoding conserved components of the pathway are indicated.
1B
strains<-c("N2", "rict-1(mg360)", "rict-1(ft7)", "sgk-1(ok538)", "akt-1(mg306)", "akt-2", "pkc-2")
foods <- "OP50"
TORC2<-read.csv(file.path(pathname, "extdata/1B_2D_rict-1_TORC2.csv"), header=TRUE) %>% dauergut::format_dauer(p.dauer = "non") #including partial/pd as non-dauer due to excess zeros.
lm <- TORC2 %>% dauer_ANOVA()
#stan
stan.glmm <- TORC2 %>% dauergut::run_dauer_stan()contrasts<-dauergut::dunnett_contrasts(lm, ref.index = 1, "genotype")
mixed<-dauergut::getStan_CIs(stan.glmm, type = "dauer")
plot.contrasts<-c("",contrasts$prange[1:6])
labels = c("WT", "rict-1(mg360)", "rict-1(ft7)", "sgk-1(ok538)", "akt-1(mg306)", "akt-2(ok393)", "pkc-2(ok328)") %>% stringr::str_wrap(width = 10)
(p<-dauergut::plot_CIs(TORC2, title='TORC2 prevents high temperature dauer formation', plot.contrasts, ypos = 1.075, type = "dauer", labels = labels))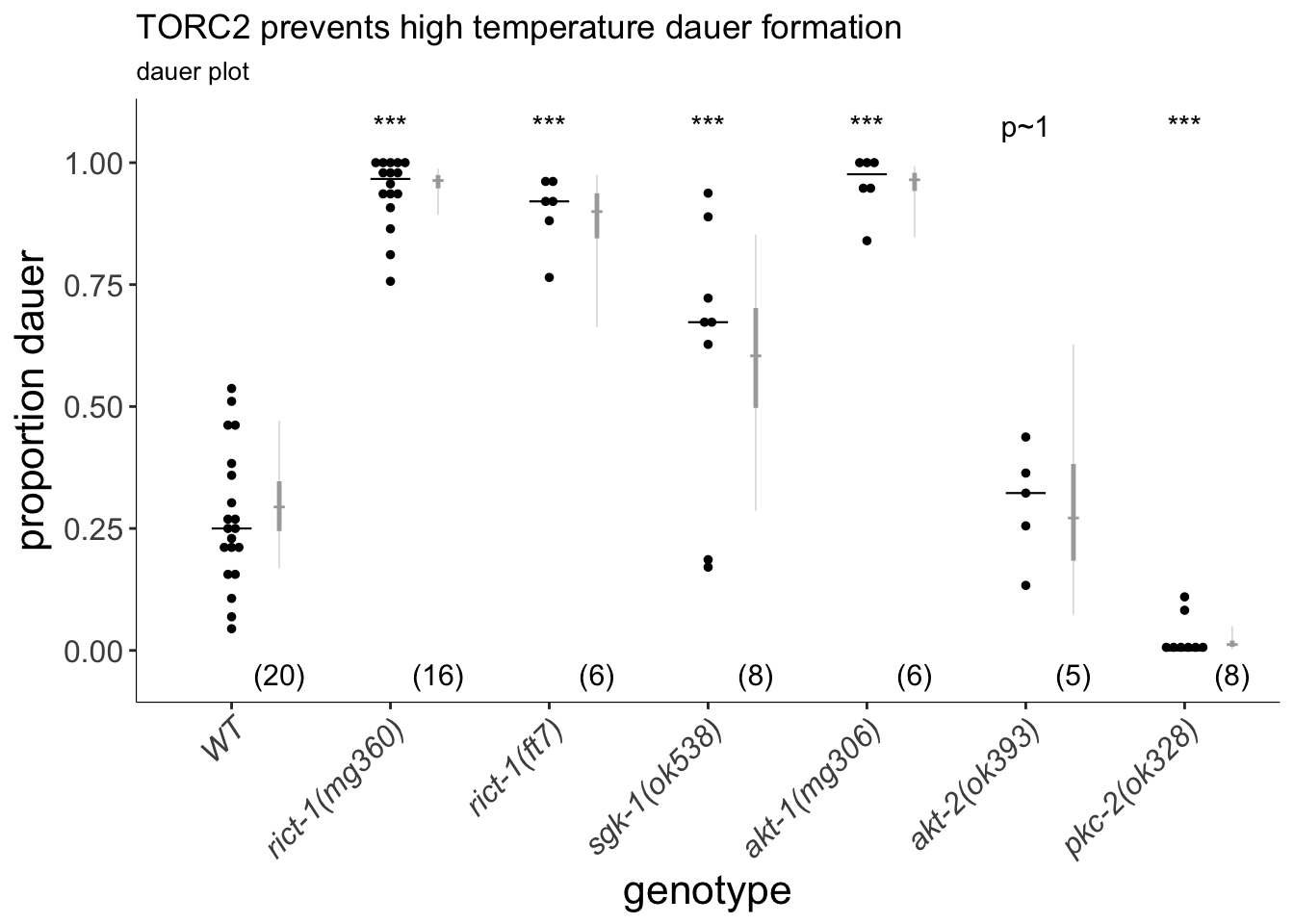
Figure 1B
TORC2 controls high temperature-induced dauer formation. Increased dauer formation occurs in TORC2-specific rict-1 mutants, as well as in mutants of TORC2 targets sgk-1 and akt-1. Plot shows proportion of dauers formed by animals of the indicated genotypes at 27°C. Each black dot indicates the proportion of dauers formed in a single assay. Arrested L3 and/or partial/post dauers are included as non-dauers due to excess arrest in rict-1(ft7). Horizontal black bar indicates median. Light gray thin and thick vertical bars at right indicate Bayesian 95% and 75% credible intervals, respectively. Numbers in parentheses below indicate the number of independent experiments with at least 25 and 9 animals each scored for non-transgenic and transgenic animals, respectively. For transgenic rescues, data are combined from 2 independent lines, with the exception of ges-1p constructs, in which one line was analyzed. *** - P<0.001 compared to wild-type (ANOVA with Dunnett-type multivariate-t adjustment for 1B and Tukey-type multivariate-t adjustment for 1C); *** - P<0.001 compared to corresponding mutant animals. P-values of differences in means relative to wild-type and corresponding mutant animals are indicated in black and red, respectively.
library(sjPlot)
sjt.lm(lm, depvar.labels = "proportion of dauers (ANOVA)", show.se = TRUE)| proportion of dauers (ANOVA) | |||||
| B | CI | std. Error | p | ||
| (Intercept) | 0.27 | 0.21 – 0.33 | 0.03 | <.001 | |
| genotype | |||||
| rict-1(mg360) | 0.67 | 0.58 – 0.76 | 0.05 | <.001 | |
| rict-1(ft7) | 0.63 | 0.50 – 0.76 | 0.06 | <.001 | |
| sgk-1(ok538) | 0.34 | 0.22 – 0.45 | 0.06 | <.001 | |
| akt-1(mg306) | 0.68 | 0.56 – 0.81 | 0.06 | <.001 | |
| akt-2 | 0.03 | -0.11 – 0.17 | 0.07 | .660 | |
| pkc-2 | -0.25 | -0.36 – -0.13 | 0.06 | <.001 | |
| Observations | 69 | ||||
| R2 / adj. R2 | .877 / .865 | ||||
| contrast | estimate | SE | df | t.ratio | p.value | prange |
|---|---|---|---|---|---|---|
| rict-1(mg360) - N2 | 0.6680907 | 0.0457575 | 62 | 14.6006908 | 0.0000000 | *** |
| rict-1(ft7) - N2 | 0.6293179 | 0.0635012 | 62 | 9.9103326 | 0.0000000 | *** |
| sgk-1(ok538) - N2 | 0.3375392 | 0.0570696 | 62 | 5.9145186 | 0.0000008 | *** |
| akt-1(mg306) - N2 | 0.6835682 | 0.0635012 | 62 | 10.7646523 | 0.0000000 | *** |
| akt-2 - N2 | 0.0301690 | 0.0682112 | 62 | 0.4422887 | 0.9975895 | p~1 |
| pkc-2 - N2 | -0.2467089 | 0.0570696 | 62 | -4.3229489 | 0.0003475 | *** |
| genotype | mean | lower.CL | upper.CL | lower.25 | upper.75 |
|---|---|---|---|---|---|
| N2 | 0.2941140 | 0.1685863 | 0.4714586 | 0.2446274 | 0.3467727 |
| rict-1(mg360) | 0.9633932 | 0.8927355 | 0.9875899 | 0.9473462 | 0.9747217 |
| rict-1(ft7) | 0.8996555 | 0.6630829 | 0.9750949 | 0.8448562 | 0.9372747 |
| sgk-1(ok538) | 0.6039456 | 0.2861732 | 0.8526753 | 0.4971906 | 0.7019556 |
| akt-1(mg306) | 0.9649093 | 0.8472189 | 0.9927722 | 0.9419640 | 0.9791813 |
| akt-2 | 0.2714936 | 0.0730530 | 0.6266923 | 0.1837610 | 0.3822809 |
| pkc-2 | 0.0119781 | 0.0023949 | 0.0490971 | 0.0073032 | 0.0201653 |
1C
strains<-c("N2","rict-1(mg360)",
"rict-1(mg360); ex[ges1]",
"rict-1(mg360); ex[gpa4]",
"rict-1(ft7)",
"rict-1(ft7); ex[ges1]",
"rict-1(ft7); ex[elt2]",
"rict-1(ft7); ex[ifb2]",
"sgk-1(ok538)",
"sgk-1(ok538); ex[ges1]",
"akt-1(mg306)",
"akt-1(mg306); ex[ges1]")
foods = "OP50"
gutresc<-read.csv(file.path(pathname, "extdata/1C_ges-1_rescue_rict_sgk.csv")) %>% dauergut::format_dauer(p.dauer = "non") %>%
mutate(allele = factor(allele, levels = c("WT","mg360","ft7","ok538","mg306")))
gutresc %<>% dplyr::mutate(adj.pct = case_when(.$pct == 0 ~ 0.01, .$pct == 1 ~ 0.99, TRUE ~ .$pct))
lm <- gutresc %>% dauer_ANOVA()
stan.glmm <- gutresc %>% dauergut::run_dauer_stan()contrasts<-dauergut::tukey_contrasts(lm, "genotype")
mixed<-stan.glmm %>% dauergut::getStan_CIs(type="dauer")
plot.contrasts<-c("",contrasts$prange[1], "", "", contrasts$prange[4], "","","", contrasts$prange[8],"",contrasts$prange[10],"")
plot.contrasts.2<-c("", "",contrasts$prange[12:13],"",contrasts$prange[39:41],"", contrasts$prange[61],"", contrasts$prange[66])
labels <- c("WT","rict-1(mg360)",
"rict-1(mg360); +ges-1p::rict-1",
"rict-1(mg360); +gpa-4p::rict-1",
"rict-1(ft7)",
"rict-1(ft7); +ges-1p::rict-1",
"rict-1(ft7); +elt-2p::rict-1",
"rict-1(ft7); +ifb-2p::rict-1",
"sgk-1(ok538)",
"sgk-1(ok538); +ges-1p::sgk-1",
"akt-1(mg306)",
"akt-1(mg306); +ges-1p::akt-1") %>% stringr::str_wrap(width=10)
(p<-dauergut::plot_CIs(gutresc, title="TORC2 components act in the intestine to regulate dauer formation", plot.contrasts, plot.contrasts.2=plot.contrasts.2, ypos = 1.075,type = "dauer", offset = 0, labels = labels))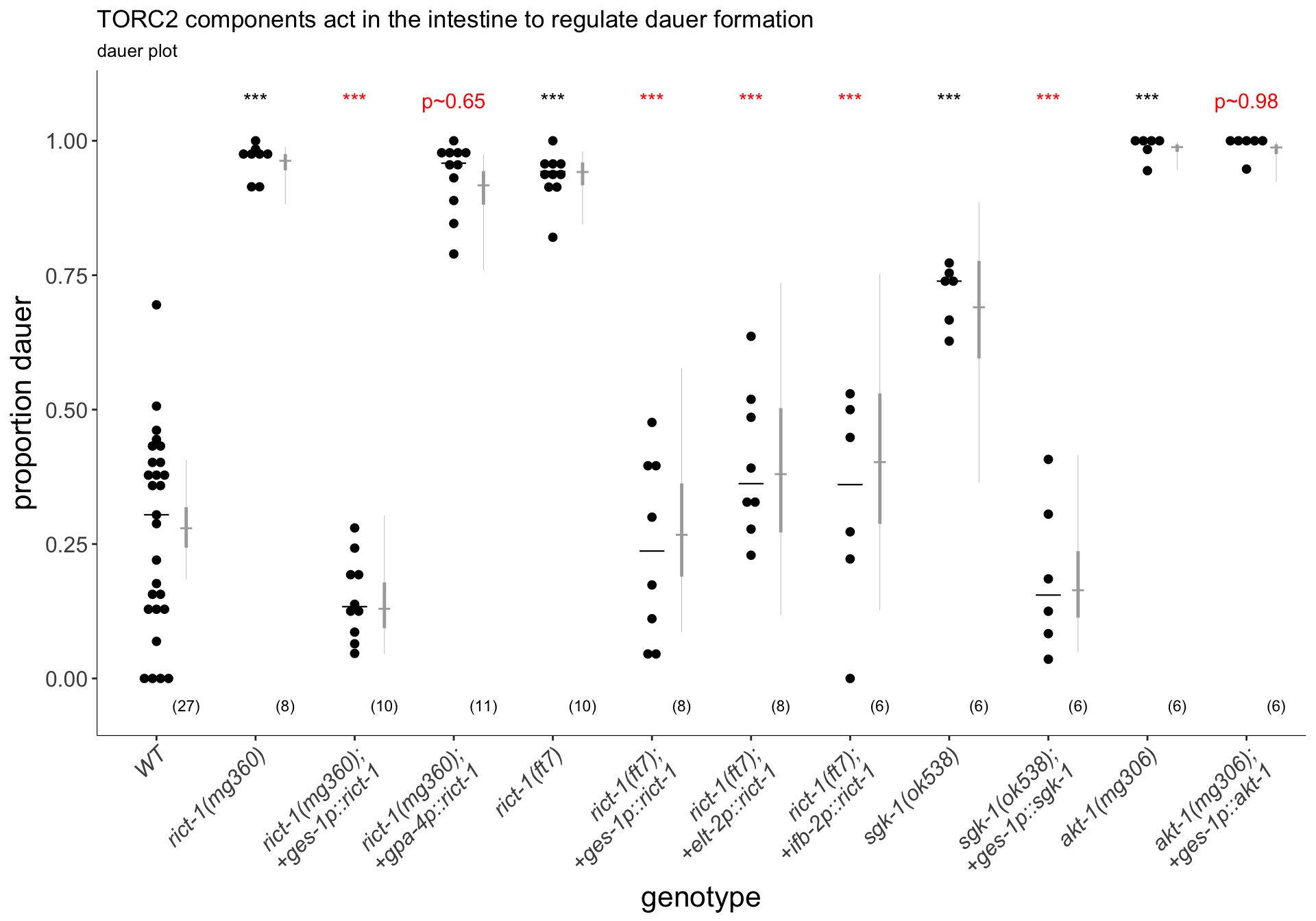
Figure 1C
mTORC2 acts in the intestine to inhibit high-temperature dauer formation. Increased dauer formation of rict-1 and sgk-1 mutants is rescued by intestinal-specific expression. akt-1 mutants were not rescued using this promoter. Dauers formed by animals of the indicated genotypes at 27°C. Each black dot indicates the proportion of dauers formed in a single assay. Horizontal black bar indicates median. Light gray thin and thick vertical bars at right indicate Bayesian 95% and 75% credible intervals, respectively. Numbers in parentheses below indicate the number of independent experiments with at least 25 and 9 animals each scored for non-transgenic and transgenic animals, respectively. For transgenic rescues, data are combined from 2 independent lines, with the exception of ges-1p constructs, in which one line was analyzed. *** - P<0.001 compared to wild-type (ANOVA with Dunnett-type multivariate-t adjustment for 1B and Tukey-type multivariate-t adjustment for 1C); *** - P<0.001 compared to corresponding mutant animals. P-values of differences in means relative to wild-type and corresponding mutant animals are indicated in black and red, respectively.
library(sjPlot)
sjt.lm(lm, depvar.labels = "proportion of dauers (ANOVA)", show.se = TRUE)| proportion of dauers (ANOVA) | |||||
| B | CI | std. Error | p | ||
| (Intercept) | 0.27 | 0.22 – 0.32 | 0.02 | <.001 | |
| genotype | |||||
| rict-1(mg360) | 0.69 | 0.59 – 0.79 | 0.05 | <.001 | |
| rict-1(mg360); ex[ges1] | -0.12 | -0.22 – -0.03 | 0.05 | .011 | |
| rict-1(mg360); ex[gpa4] | 0.66 | 0.57 – 0.75 | 0.05 | <.001 | |
| rict-1(ft7) | 0.66 | 0.57 – 0.75 | 0.05 | <.001 | |
| rict-1(ft7); ex[ges1] | -0.03 | -0.13 – 0.07 | 0.05 | .556 | |
| rict-1(ft7); ex[elt2] | 0.13 | 0.02 – 0.23 | 0.05 | .017 | |
| rict-1(ft7); ex[ifb2] | 0.06 | -0.06 – 0.17 | 0.06 | .344 | |
| sgk-1(ok538) | 0.44 | 0.33 – 0.56 | 0.06 | <.001 | |
| sgk-1(ok538); ex[ges1] | -0.08 | -0.20 – 0.03 | 0.06 | .155 | |
| akt-1(mg306) | 0.71 | 0.60 – 0.83 | 0.06 | <.001 | |
| akt-1(mg306); ex[ges1] | 0.72 | 0.60 – 0.83 | 0.06 | <.001 | |
| Observations | 112 | ||||
| R2 / adj. R2 | .884 / .871 | ||||
| contrast | estimate | SE | df | t.ratio | p.value | prange |
|---|---|---|---|---|---|---|
| N2 - rict-1(mg360) | -0.6909880 | 0.0518355 | 100 | -13.3303901 | 0.0000000 | *** |
| N2 - rict-1(mg360); ex[ges1] | 0.1241772 | 0.0476694 | 100 | 2.6049685 | 0.0155310 | p~0.016 |
| N2 - rict-1(mg360); ex[gpa4] | -0.6605064 | 0.0460611 | 100 | -14.3397941 | 0.0000000 | *** |
| N2 - rict-1(ft7) | -0.6599220 | 0.0476694 | 100 | -13.8437300 | 0.0000000 | *** |
| N2 - rict-1(ft7); ex[ges1] | 0.0305997 | 0.0518355 | 100 | 0.5903222 | 0.6119370 | p~0.61 |
| N2 - rict-1(ft7); ex[elt2] | -0.1258822 | 0.0518355 | 100 | -2.4284924 | 0.0237990 | p~0.024 |
| N2 - rict-1(ft7); ex[ifb2] | -0.0552211 | 0.0581192 | 100 | -0.9501352 | 0.4287936 | p~0.43 |
| N2 - sgk-1(ok538) | -0.4428609 | 0.0581192 | 100 | -7.6198664 | 0.0000000 | *** |
| N2 - sgk-1(ok538); ex[ges1] | 0.0831857 | 0.0581192 | 100 | 1.4312943 | 0.2052121 | p~0.21 |
| N2 - akt-1(mg306) | -0.7145009 | 0.0581192 | 100 | -12.2937048 | 0.0000000 | *** |
| N2 - akt-1(mg306); ex[ges1] | -0.7176764 | 0.0581192 | 100 | -12.3483425 | 0.0000000 | *** |
| rict-1(mg360) - rict-1(mg360); ex[ges1] | 0.8151652 | 0.0610818 | 100 | 13.3454703 | 0.0000000 | *** |
| rict-1(mg360) - rict-1(mg360); ex[gpa4] | 0.0304817 | 0.0598351 | 100 | 0.5094278 | 0.6516490 | p~0.65 |
| rict-1(mg360) - rict-1(ft7) | 0.0310660 | 0.0610818 | 100 | 0.5085968 | 0.6516490 | p~0.65 |
| rict-1(mg360) - rict-1(ft7); ex[ges1] | 0.7215877 | 0.0643859 | 100 | 11.2072381 | 0.0000000 | *** |
| rict-1(mg360) - rict-1(ft7); ex[elt2] | 0.5651058 | 0.0643859 | 100 | 8.7768615 | 0.0000000 | *** |
| rict-1(mg360) - rict-1(ft7); ex[ifb2] | 0.6357669 | 0.0695447 | 100 | 9.1418477 | 0.0000000 | *** |
| rict-1(mg360) - sgk-1(ok538) | 0.2481271 | 0.0695447 | 100 | 3.5678807 | 0.0009148 | *** |
| rict-1(mg360) - sgk-1(ok538); ex[ges1] | 0.7741738 | 0.0695447 | 100 | 11.1320344 | 0.0000000 | *** |
| rict-1(mg360) - akt-1(mg306) | -0.0235128 | 0.0695447 | 100 | -0.3380969 | 0.7589989 | p~0.76 |
| rict-1(mg360) - akt-1(mg306); ex[ges1] | -0.0266883 | 0.0695447 | 100 | -0.3837582 | 0.7353996 | p~0.74 |
| rict-1(mg360); ex[ges1] - rict-1(mg360); ex[gpa4] | -0.7846836 | 0.0562644 | 100 | -13.9463561 | 0.0000000 | *** |
| rict-1(mg360); ex[ges1] - rict-1(ft7) | -0.7840992 | 0.0575885 | 100 | -13.6155604 | 0.0000000 | *** |
| rict-1(mg360); ex[ges1] - rict-1(ft7); ex[ges1] | -0.0935776 | 0.0610818 | 100 | -1.5320041 | 0.1733227 | p~0.173 |
| rict-1(mg360); ex[ges1] - rict-1(ft7); ex[elt2] | -0.2500595 | 0.0610818 | 100 | -4.0938460 | 0.0001537 | *** |
| rict-1(mg360); ex[ges1] - rict-1(ft7); ex[ifb2] | -0.1793984 | 0.0664974 | 100 | -2.6978240 | 0.0122898 | p~0.012 |
| rict-1(mg360); ex[ges1] - sgk-1(ok538) | -0.5670381 | 0.0664974 | 100 | -8.5272183 | 0.0000000 | *** |
| rict-1(mg360); ex[ges1] - sgk-1(ok538); ex[ges1] | -0.0409915 | 0.0664974 | 100 | -0.6164370 | 0.6029569 | p~0.6 |
| rict-1(mg360); ex[ges1] - akt-1(mg306) | -0.8386781 | 0.0664974 | 100 | -12.6121875 | 0.0000000 | *** |
| rict-1(mg360); ex[ges1] - akt-1(mg306); ex[ges1] | -0.8418536 | 0.0664974 | 100 | -12.6599413 | 0.0000000 | *** |
| rict-1(mg360); ex[gpa4] - rict-1(ft7) | 0.0005843 | 0.0562644 | 100 | 0.0103857 | 0.9917342 | p~1 |
| rict-1(mg360); ex[gpa4] - rict-1(ft7); ex[ges1] | 0.6911060 | 0.0598351 | 100 | 11.5501788 | 0.0000000 | *** |
| rict-1(mg360); ex[gpa4] - rict-1(ft7); ex[elt2] | 0.5346241 | 0.0598351 | 100 | 8.9349594 | 0.0000000 | *** |
| rict-1(mg360); ex[gpa4] - rict-1(ft7); ex[ifb2] | 0.6052852 | 0.0653541 | 100 | 9.2616222 | 0.0000000 | *** |
| rict-1(mg360); ex[gpa4] - sgk-1(ok538) | 0.2176455 | 0.0653541 | 100 | 3.3302483 | 0.0019578 | ** |
| rict-1(mg360); ex[gpa4] - sgk-1(ok538); ex[ges1] | 0.7436921 | 0.0653541 | 100 | 11.3794210 | 0.0000000 | *** |
| rict-1(mg360); ex[gpa4] - akt-1(mg306) | -0.0539945 | 0.0653541 | 100 | -0.8261836 | 0.4790578 | p~0.48 |
| rict-1(mg360); ex[gpa4] - akt-1(mg306); ex[ges1] | -0.0571700 | 0.0653541 | 100 | -0.8747727 | 0.4646272 | p~0.46 |
| rict-1(ft7) - rict-1(ft7); ex[ges1] | 0.6905217 | 0.0610818 | 100 | 11.3048694 | 0.0000000 | *** |
| rict-1(ft7) - rict-1(ft7); ex[elt2] | 0.5340398 | 0.0610818 | 100 | 8.7430275 | 0.0000000 | *** |
| rict-1(ft7) - rict-1(ft7); ex[ifb2] | 0.6047009 | 0.0664974 | 100 | 9.0935972 | 0.0000000 | *** |
| rict-1(ft7) - sgk-1(ok538) | 0.2170611 | 0.0664974 | 100 | 3.2642030 | 0.0023622 | ** |
| rict-1(ft7) - sgk-1(ok538); ex[ges1] | 0.7431078 | 0.0664974 | 100 | 11.1749842 | 0.0000000 | *** |
| rict-1(ft7) - akt-1(mg306) | -0.0545788 | 0.0664974 | 100 | -0.8207663 | 0.4790578 | p~0.48 |
| rict-1(ft7) - akt-1(mg306); ex[ges1] | -0.0577544 | 0.0664974 | 100 | -0.8685200 | 0.4646272 | p~0.46 |
| rict-1(ft7); ex[ges1] - rict-1(ft7); ex[elt2] | -0.1564819 | 0.0643859 | 100 | -2.4303767 | 0.0237990 | p~0.024 |
| rict-1(ft7); ex[ges1] - rict-1(ft7); ex[ifb2] | -0.0858208 | 0.0695447 | 100 | -1.2340386 | 0.2848105 | p~0.28 |
| rict-1(ft7); ex[ges1] - sgk-1(ok538) | -0.4734606 | 0.0695447 | 100 | -6.8080056 | 0.0000000 | *** |
| rict-1(ft7); ex[ges1] - sgk-1(ok538); ex[ges1] | 0.0525861 | 0.0695447 | 100 | 0.7561481 | 0.5135916 | p~0.51 |
| rict-1(ft7); ex[ges1] - akt-1(mg306) | -0.7451005 | 0.0695447 | 100 | -10.7139833 | 0.0000000 | *** |
| rict-1(ft7); ex[ges1] - akt-1(mg306); ex[ges1] | -0.7482760 | 0.0695447 | 100 | -10.7596446 | 0.0000000 | *** |
| rict-1(ft7); ex[elt2] - rict-1(ft7); ex[ifb2] | 0.0706611 | 0.0695447 | 100 | 1.0160530 | 0.3960713 | p~0.4 |
| rict-1(ft7); ex[elt2] - sgk-1(ok538) | -0.3169787 | 0.0695447 | 100 | -4.5579140 | 0.0000269 | *** |
| rict-1(ft7); ex[elt2] - sgk-1(ok538); ex[ges1] | 0.2090680 | 0.0695447 | 100 | 3.0062396 | 0.0051334 | ** |
| rict-1(ft7); ex[elt2] - akt-1(mg306) | -0.5886186 | 0.0695447 | 100 | -8.4638917 | 0.0000000 | *** |
| rict-1(ft7); ex[elt2] - akt-1(mg306); ex[ges1] | -0.5917941 | 0.0695447 | 100 | -8.5095530 | 0.0000000 | *** |
| rict-1(ft7); ex[ifb2] - sgk-1(ok538) | -0.3876397 | 0.0743464 | 100 | -5.2139687 | 0.0000019 | *** |
| rict-1(ft7); ex[ifb2] - sgk-1(ok538); ex[ges1] | 0.1384069 | 0.0743464 | 100 | 1.8616491 | 0.0901854 | p~0.09 |
| rict-1(ft7); ex[ifb2] - akt-1(mg306) | -0.6592797 | 0.0743464 | 100 | -8.8676763 | 0.0000000 | *** |
| rict-1(ft7); ex[ifb2] - akt-1(mg306); ex[ges1] | -0.6624552 | 0.0743464 | 100 | -8.9103885 | 0.0000000 | *** |
| sgk-1(ok538) - sgk-1(ok538); ex[ges1] | 0.5260466 | 0.0743464 | 100 | 7.0756178 | 0.0000000 | *** |
| sgk-1(ok538) - akt-1(mg306) | -0.2716400 | 0.0743464 | 100 | -3.6537076 | 0.0007003 | *** |
| sgk-1(ok538) - akt-1(mg306); ex[ges1] | -0.2748155 | 0.0743464 | 100 | -3.6964198 | 0.0006203 | *** |
| sgk-1(ok538); ex[ges1] - akt-1(mg306) | -0.7976866 | 0.0743464 | 100 | -10.7293254 | 0.0000000 | *** |
| sgk-1(ok538); ex[ges1] - akt-1(mg306); ex[ges1] | -0.8008621 | 0.0743464 | 100 | -10.7720376 | 0.0000000 | *** |
| akt-1(mg306) - akt-1(mg306); ex[ges1] | -0.0031755 | 0.0743464 | 100 | -0.0427122 | 0.9808779 | p~0.98 |
| genotype | mean | lower.CL | upper.CL | lower.25 | upper.75 |
|---|---|---|---|---|---|
| N2 | 0.2793225 | 0.1833459 | 0.4066347 | 0.2431513 | 0.3185055 |
| rict-1(mg360) | 0.9627715 | 0.8815987 | 0.9885193 | 0.9450084 | 0.9752851 |
| rict-1(mg360); ex[ges1] | 0.1296133 | 0.0457332 | 0.3034522 | 0.0935004 | 0.1785384 |
| rict-1(mg360); ex[gpa4] | 0.9170733 | 0.7598877 | 0.9740302 | 0.8811744 | 0.9436661 |
| rict-1(ft7) | 0.9420074 | 0.8443246 | 0.9802061 | 0.9171760 | 0.9596365 |
| rict-1(ft7); ex[ges1] | 0.2671974 | 0.0875506 | 0.5767962 | 0.1894259 | 0.3626919 |
| rict-1(ft7); ex[elt2] | 0.3800157 | 0.1172738 | 0.7357145 | 0.2715965 | 0.5027534 |
| rict-1(ft7); ex[ifb2] | 0.4023557 | 0.1275289 | 0.7527617 | 0.2873637 | 0.5301470 |
| sgk-1(ok538) | 0.6903346 | 0.3638554 | 0.8856750 | 0.5950865 | 0.7766973 |
| sgk-1(ok538); ex[ges1] | 0.1641085 | 0.0480079 | 0.4154684 | 0.1131033 | 0.2367389 |
| akt-1(mg306) | 0.9881795 | 0.9461830 | 0.9977387 | 0.9794053 | 0.9930754 |
| akt-1(mg306); ex[ges1] | 0.9875661 | 0.9235693 | 0.9985239 | 0.9752865 | 0.9933613 |